Unveiling the Catalytic Mechanism and Conformational Dynamics of Guinea Pig l-Asparaginase Type 1 for Leukemia Drug Design
Abstract
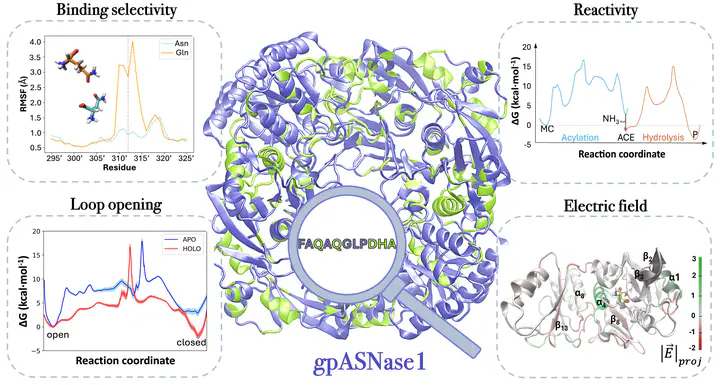
In this study, we present a computational analysis of the catalytic properties of guinea pig asparaginase type 1 (or gpASNase1), an enzyme of mammalian origin that offers a potential alternative for the treatment of acute lymphoblastic leukemia. This enzyme transforms asparagine into aspartate, depriving leukemia cells of this essential amino acid. A combination of molecular dynamics simulations, free energy calculations, and mechanistic insights based on quantum mechanics/molecular mechanics hybrid approaches was used to identify those residues contributing to the catalytic cycle of the enzyme. We dissected the contribution of enzymatic residues to substrate binding and selectivity, showing why this ASNase can degrade asparagine but not glutamine. We also studied the conformational dynamics of the enzymatic loop closing the active site, demonstrating that the substrate binding favors the closed state. The catalytic reaction mechanisms, composed of two stages, acylation and hydrolysis, were explored as well. The rate-limiting step presents a free energy barrier close to the experimental estimation and corresponds to the nucleophilic attack of enzymatic Thr19 on the carbonyl carbon atom of the substrate. Analysis of the electric field created by the protein sheds light on the role of certain residues and structural motifs in stabilizing the reaction transition state. The conclusions of this analysis are useful for rationalizing the properties of chimeras derived from gpASNase1 and predicting additional residue positions, where mutations could enhance substrate binding and loop dynamics. The results of this study enhance the understanding of gpASNase1, offering valuable insights into rational mutations and enzyme engineering for the treatment of leukemia.